Julia Brasch
Assistant Professor of Biochemistry
Synaptic Organization, Cell-Cell Adhesion, Structural Biology, Electron Cryo-tomography, cryo-EM, X-ray Crystallography, Biophysics

Molecular Biology Program
Biological Chemistry Program
Education
B.S., M.S. Gottfried Wilhelm Leibniz University of Hannover
Ph.D. Gottfried Wilhelm Leibniz University of Hannover
Research
Neurons in the brain form complex neural circuits by connecting to each other through highly specialized junctions called synapses. A molecular logic underlies the formation, establishment and properties of each of these synapses and is likely driven by synaptic cell adhesion molecules. In the lab we aim to understand the protein complexes formed at these junctions and how they assemble and arrange in respect to each other in the synaptic cleft with the overall aim to understand the extracellular architecture of the synapse. We use a combination of cryo-electron microscopy, electron cryo-tomography and biophysical techniques to understand their interactions at a molecular level and relate concepts determined in vitro back to biology. Our approach identifies the building blocks of the synapse and will help to understand how these crucial elements of our nervous system are assembled. Join us in this fascinating line of research using cutting edge techniques!
References (Selected Publications)
- Harrison OJ*, Brasch J*, Katsamba PS, Ahlsen G, Noble AJ, Dan H, Sampogna RV, Potter CS, Carragher B, Honig B, Shapiro L. Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered δ-Protocadherins. Cell Rep. 2020 Feb 25;30(8):2655-2671.e7. doi: 10.1016/j.celrep.2020.02.003. PMID: 32101743; PMCID: PMC7082078
- Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, Berger B. Positive- unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods. 2019 Nov;16(11):1153-1160. doi: 10.1038/s41592-019-0575-8. Epub 2019 Oct 7. PMID: 31591578; PMCID: PMC6858545
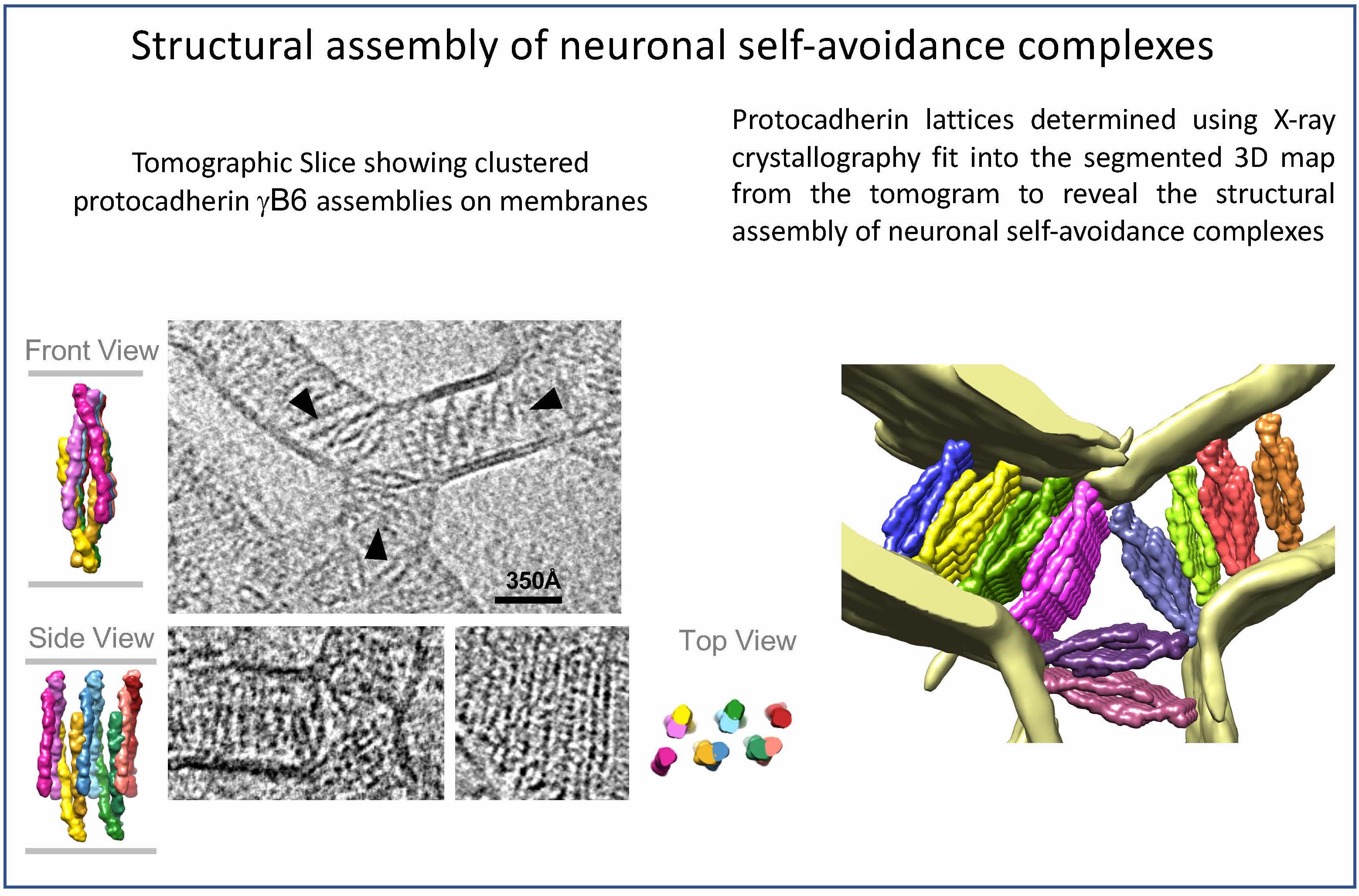
- Brasch J*, Goodman KM*, Noble AJ, Rapp M, Mannepalli S, Bahna F, Dandey VP, Bepler T, Berger B, Maniatis T, Potter CS, Carragher B, Honig B, Shapiro L. Visualization of clustered protocadherin neuronal self-recognition complexes. Nature. 2019 May;569(7755):280-283. doi: 10.1038/s41586-019-1089-3. Epub 2019 Apr 10. PMID: 30971825; PMCID: PMC6736547
- Noble AJ, Dandey VP, Wei H, Brasch J, Chase J, Acharya P, Tan YZ, Zhang Z, Kim LY, Scapin G, Rapp M, Eng ET, Rice WJ, Cheng A, Negro CJ, Shapiro L, Kwong PD, Jeruzalmi D, des Georges A, Potter CS, Carragher B. Routine single particle CryoEM sample and grid characterization by tomography. Elife. 2018 May 29;7:e34257. doi: 10.7554/eLife.34257. PMID: 29809143; PMCID: PMC5999397
- Brasch J, Katsamba PS, Harrison OJ, Ahlsén G, Troyanovsky RB, Indra I, Kaczynska A, Kaeser B, Troyanovsky S, Honig B, Shapiro L. Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior. Cell Rep. 2018 May 8;23(6):1840-1852. doi: 10.1016/j.celrep.2018.04.012. PMID: 29742438; PMCID: PMC6029887
- Harrison OJ*, Brasch J*, Lasso G, Katsamba PS, Ahlsen G, Honig B, Shapiro L. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):7160-5. doi: 10.1073/pnas.1606272113. Epub 2016 Jun 13. PMID: 27298358; PMCID: PMC4932976
