Raphael Franzini
Associate Professor of Medicinal Chemistry
Drug Development, Chemical Probes, Imaging Agents

Biological Chemistry Program
Education
M.S. Swiss Federal Institute of Technology, Lausanne, Switzerland
Ph.D. Stanford University
Research
Our research group aims to pioneer innovative chemistry for groundbreaking therapeutics. As a Medicinal Chemistry group, we harness organic synthesis, bioconjugation, affinity selections, imaging, and computation for the purpose of drug delivery. Our research activities prioritize the training and growth of students and scientists, fostering a scientific environment that drives cutting-edge innovations and personal advancement. The two main research areas center around DNA-encoded libraries for drug discovery and localized delivery of drugs using bioorthogonal chemistry.
Fishing for Drugs with DNA-encoded Libraries
Drug discovery is an arduous process. Wouldn’t it be nice to be able to simply fish out drug candidates from a pool of molecules? That’s exactly what DNA-encoded libraires aim to accomplish. DNA-encoded libraries consist of collections of small molecules that have an appended DNA-tag that encodes the structure of the molecule. With DNA-encoded libraries, one can screen millions to billions of molecules in a couple of hours and on top of that obtain valuable structure-activity information. Unsurprisingly, DNA-encoded libraries are increasingly used in the pharmaceutical industry, and the Franzini lab is one of few training environments where scientists can get exposure to all aspects of the technology. Our interest with DNA-encoded libraries is to advance the technology while also use it for identifying drug leads. Of particular interest are approaches that combine DNA-encoded libraries with computational approaches. This can involve using the DNA-encoded library data as input data to computationally predict lead molecules. Alternatively, we are interested in using computational methods to aid the design of DNA-encoded libraries to tackle undruggable targets.
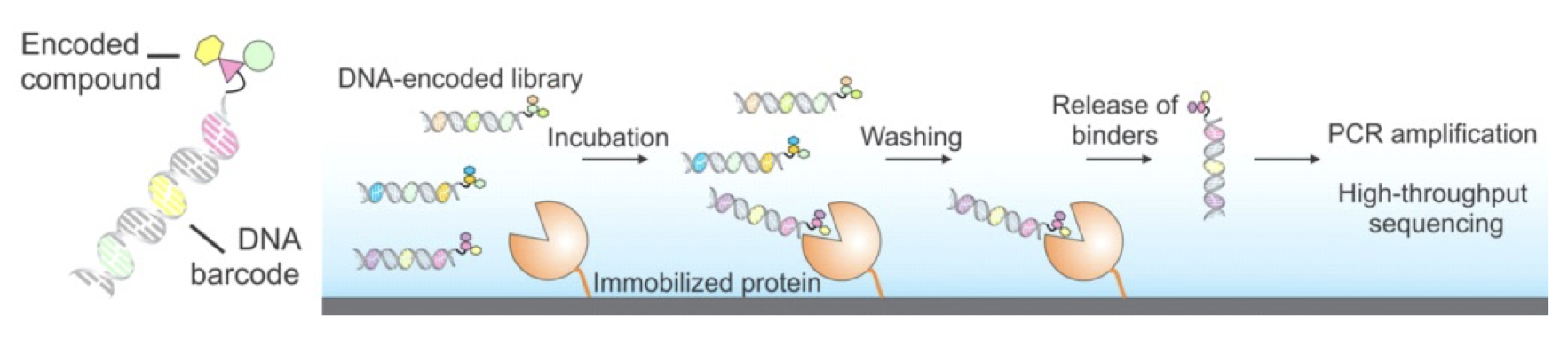
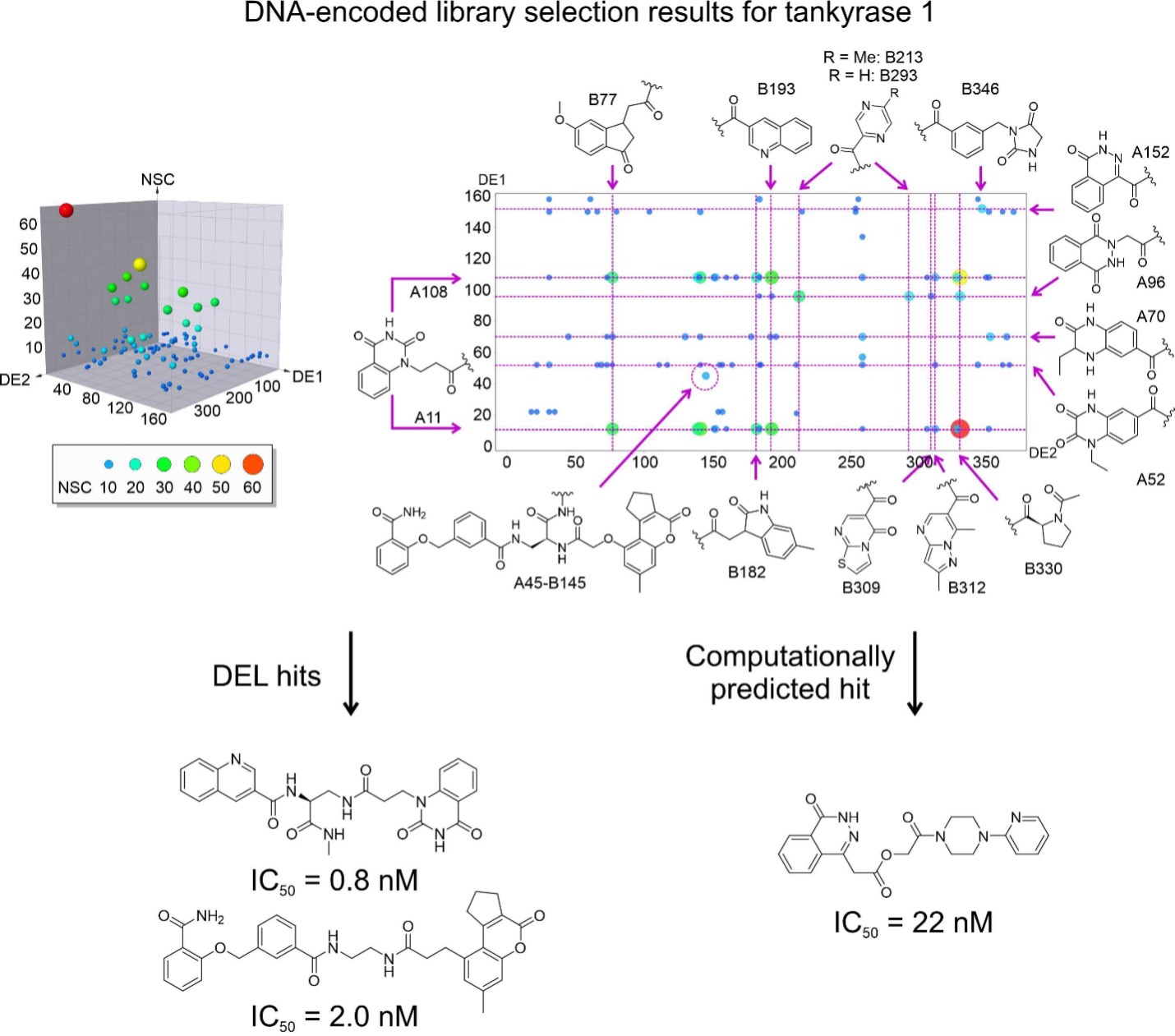
Figure 1. DNA-encoded libraries for drug discovery. Upper figure: Concept of DNA-encoded library screening by affinity selection. Lower figure: Discovery of highly potent inhibitors from DNA-encoded libraries as an example.
Using Chemistry to Deliver Drugs to Where They are Needed
Some diseases, and in particular solid tumors, are localized, and delivering a drug specifically to the site of disease is a proven strategy to enhance therapeutic outcome while minimizing side effects. We are combining basic organic chemistry research with drug delivery approaches to pioneer new types of localized drugs. In one approach, we develop new linker chemistry that allows us to control the distribution and release of drugs. In a second approach, we are developing bioorthogonal drug release schemes that allows us to deliver the drug where and when it is needed. For this purpose, we have developed and are developing new drug delivery chemistry. In particular, we have successfully explored the use of the isocyano group to establish drug-release chemistry. The first drugs based on bioorthogonal chemistry have recently entered clinical trials, and this will be an exciting research field to be in for the years to come.
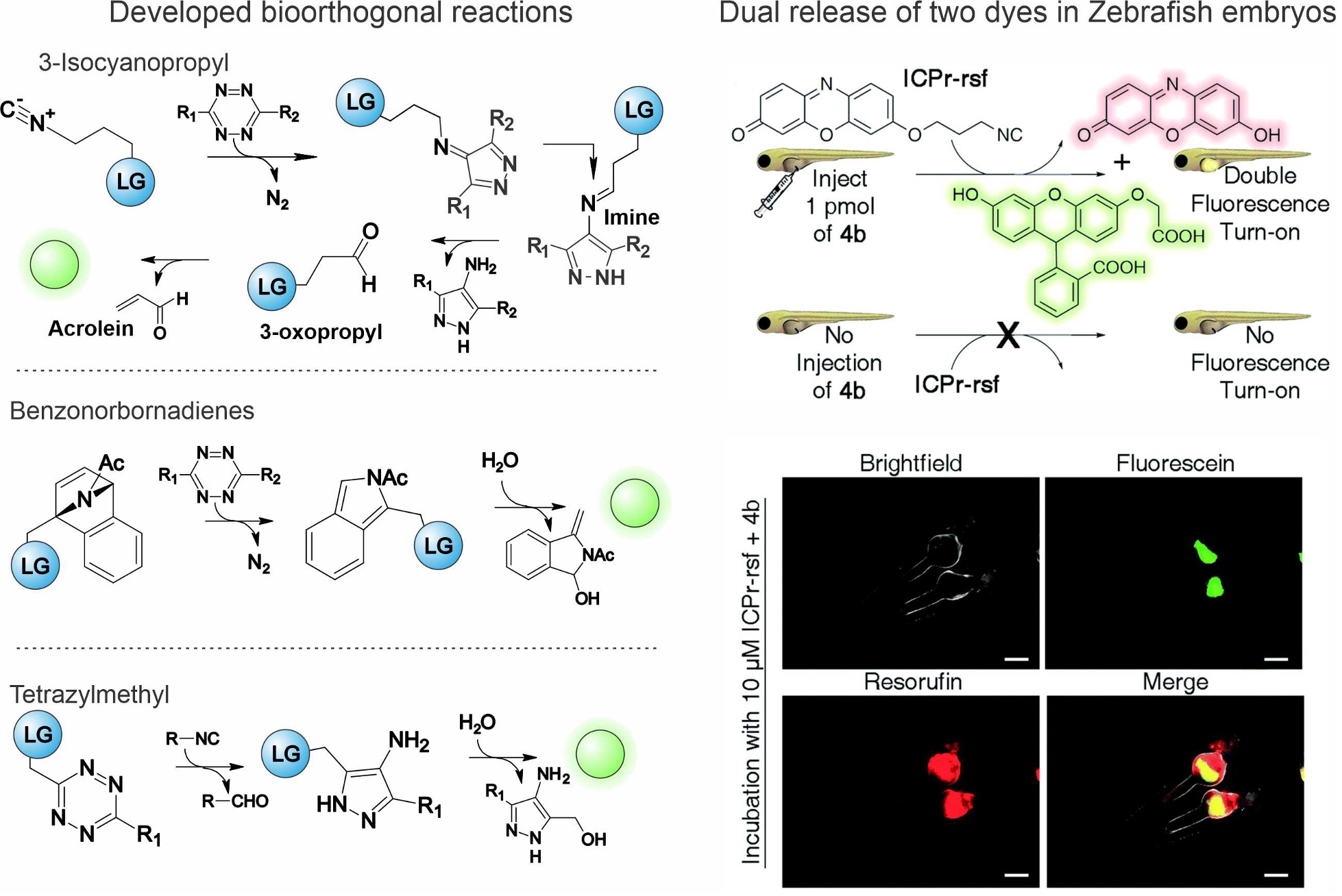
Figure 2. Bioorthogonal release chemistry developed in our lab and demonstration experiment releasing two dyes simultaneously in Zebrafish embryos.
References (Selected Publications)
DNA-encoded Libraries:
- Montoya et al. Combining pharmacophore models derived from DNA-encoded chemical libraries with structure-based exploration to predict Tankyrase 1 inhibitors. Eur. J. Med. Chem., 2023, 246, 114980. (https://www.sciencedirect.com/science/article/pii/S0223523422008820)
- Lemke et al. Integrating DNA-encoded chemical libraries with virtual combinatorial library screening: Optimizing a PARP10 inhibitor. Bioorg. Med. Chem. Letts.,2020, 30, 127464. (https://www.sciencedirect.com/science/article/pii/S0960894X20305758)
- H. Yuen et al. A Focused DNA-Encoded Chemical Library for the Discovery of Inhibitors of NAD+-Dependent Enzymes. J. Am. Chem. Soc., 2019, 141, 5169. (https://pubs.acs.org/doi/full/10.1021/jacs.8b08039)
Bioorthogonal Drug-release Chemistry:
- Tu et al. Isonitrile-responsive and bioorthogonally removable tetrazine protecting groups. Chem. Sci., 2020, 11, 169. (https://pubs.rsc.org/en/content/articlehtml/2019/sc/c9sc04649f)
- Tu et al. Stable, reactive, and orthogonal tetrazines: dispersion forces promote the cycloaddition with isonitriles. Angew. Chem. Int. Ed., 2019, 58, 9043. (https://onlinelibrary.wiley.com/doi/full/10.1002/anie.201903877)
- Tu et al. Bioorthogonal removal of 3-isocyanopropyl groups enables the controlled release of fluorophores and drugs in vivo. J. Am. Chem. Soc., 2018, 140, 8410. (https://pubs.acs.org/doi/full/10.1021/jacs.8b05093)
- Xu et al. Rapid and efficient tetrazine-induced drug release from highly stable benzonorbornadiene derivatives, Chem.Commun., 2017, 53, 6271. (http://pubs.rsc.org/en/Content/ArticleLanding/2017/CC/C7CC03477F#!divAbstract)
