Ming Hammond
Professor of Chemistry
Biosensors, RNA, Fluorescence Imaging, Bacterial and Immune Cell Signaling

Biological Chemistry Program
Education
B.S. California Institute of Technology
Ph.D. University of California, Berkeley
Research
Illuminating the Single-Cell Biology and Function of Chemical Signals
The Hammond lab has a dual focus on engineering nucleic acids as programmable tools
for molecular imaging and gene control, and on understanding the chemistry and biology
of cyclic dinucleotides as signaling molecules in bacteria and mammalian cells.
Molecular Imaging: RNA-based fluorescent biosensors

We are one of the first labs to develop fluorescent biosensors made of RNA for live cell imaging of enzyme activity. These sensors are designed by combining a riboswitch domain, which is an RNA that changes conformation upon binding a small molecule ligand, and a fluorophore-binding domain. Ligand selectivity is dictated by the riboswitch domain and can even be reprogrammed with single nucleotide changes.
Our lab has established allosteric stem design rules, novel designs, and optimization strategies for making RNA-based fluorescent biosensors from a variety of riboswitch folds. We have made biosensors with best-in-class sensitivity (sub-nanomolar limit of detection), selectivity (better than commercial monoclonal antibodies), and in vivo brightness (143% brighter than parent fluorophore-binding aptamer). We have applied these biosensors to visualize enzyme activity in both gram positive and gram negative bacteria, under aerobic and anaerobic conditions, via fluorescence microscopy and flow cytometry methods. We have 'watched' the chemical inhibition of an enzyme involved in biosynthesis of a quorum signaling molecule in live bacterial cells. We recently showed that the micronutrient zinc turns off a signaling enzyme that controls biofilm formation in E. coli, which helps to sensitize this common foodborne pathogen to antibiotics (Future of Biochemistry).
Biosensor Papers: JACS 2013a, JACS 2015, JACS 2016, NAR 2016, Cell Chem Biol 2016, Biochem 2018, Methods 2018, Biopolymers 2020, ACS Synth Bio 2021
Other Riboswitch-Based Methods Papers: JACS 2013b, Chem Biol 2013, Anal Chem 2014, RNA Biol 2015
Reviews: Methods Enz 2015, Methods Mol Biol 2015, Methods Mol Biol 2017, Annu Rev Biochem 2017, Microbiol Spec 2018, Regulation with RNA in Bacteria and Archaea 2019, Curr Opin Biotech 2020, Methods Enz 2020, Curr Opin Chem Biol 2020, Methods Mol Bio 2021
Signaling: Cyclic dinucleotides in bacteria and mammalian cells
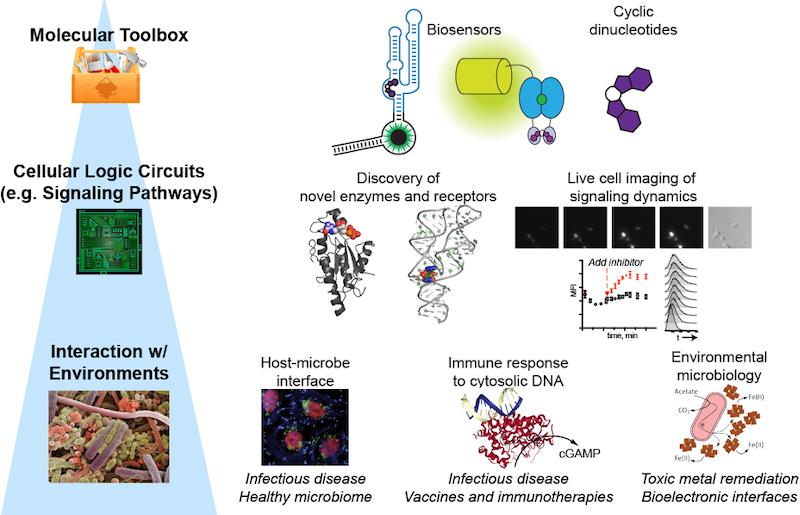
Cyclic dinucleotides are an emerging class of intracellular signaling molecules with diverse roles in bacteria and with newfound roles in innate immunity in mammalian cells. As second messengers, they are produced transiently in response to environmental stimuli and act inside the cell to change gene expression, physiology, and behavior. Given that three out of the four known cyclic dinucleotides were discovered in the past decade, much remains to be explored about their chemistry and biology. To study these chemical signals, our lab has developed fluorescent biosensors for each of the known cyclic dinucleotides. We also recently developed the first bioluminescent sensor for measuring cyclic dinucleotides in complex settings (Nat Chem Biol Research Highlight, JGI Science Highlight).
(i) Cyclic AMP-GMP (Bacterial cGAMP) signaling: Using an in vitro biosensor assay, we discovered a cyclic AMP-GMP (cGAMP)-sensing riboswitch class, which was highlighted by Science Signaling as a Signaling Breakthrough of the Year. These newfound cGAMP riboswitches were identified in Geobacter bacteria and predicted to control these bacteria's unique ability to relay electrons outside the cell. By screening for enzyme activity using an in vivo biosensor assay, we subsequently discovered a G. sulfurreducens GGDEF enzyme that makes cGAMP, which is the founding member of Hypr GGDEF enzymes. This discovery is significant because for almost 30 years GGDEF enzymes were considered synonymous with the classical cyclic di-GMP signaling network; our result provides the first evidence that this enzyme class can make alternative cyclic dinucleotides to cyclic di-GMP (Berkeley news story). We are continuing to identify molecular components of the cGAMP signaling pathway and to understand its role in how bacteria like Geobacter sense and adapt to different kinds of surfaces (eLife digest story).
(ii) Mammalian cGAMP signaling: The enzyme cyclic GMP-AMP synthase (cGAS) was discovered to be the innate immune sensor for cytosolic DNA, which may result from microbial infection or other pathophysiological conditions. Upon binding double-stranded DNA, cGAS produces the signaling molecule cGAMP, which activates the receptor protein, stimulator of interferon genes (STING). In collaboration with Russell Vance (UC Berkeley), we proved the chemical structure of cGAMP had a noncanonical 2'-5' linkage by NMR. We recently developed a biosensor-based platereader assay to quantitate cGAMP levels in DNA-stimulated mammalian cell lysates.
(iii) Other cyclic dinucleotide signaling: We have analyzed the activity of cyclic di-GMP and cyclic di-AMP signaling enzymes using in vivo biosensor assays. For example, we have demonstrated that archaeal enzymes produce cyclic di-AMP. This result completed the set of experimental evidence that cyclic dinucleotide signaling extends to all three domains of life.
Bacterial cGAMP Papers: PNAS 2015, Cell Reports 2015, PNAS 2016, eLIFE 2019, bioRχiv 2019, Microbial Cyclic Di-Nucleotide Signaling 2020, Molecular Microbiology 2020
Mammalian cGAMP Papers: Cell Reports 2013, Cell Chem Biol 2016, ChemBioChem 2020
Other CDN Biosensor Papers: JACS 2013a, JACS 2015, NAR 2016, ACS Chem Bio 2018, ACS Chem Biol 2020
References
- Mumbleau, M.M., Meyer, M.R., Hammond, M.C. "Determination of in Vitro and in Vivo turn-on kinetics for Fluorogenic RNA aptamers" JOVE (2022) Invited Paper - Under Review.
- Banuelos Jara, B. and Hammond, M.C. "Functions and applications of riboswitches" Handbook of Chemical Biology of Nucleic Acids (2022) Invited Chapter - Under Review.
- Mukkayyan, N., Poon, R., Sander, P.N., Lai, L-Y., Zubair-Nizami, Z., Hammond, M.C., Chatterjee, S.S. "In-vivo detection of cyclic-di-AMP in Staphylococcus aureus" bioRxiv (2022) 10, 1101 -Under Review.
- Lowry, R.C., Hallberg, Z.F., Till, R., Simons, T.J., Nottingham, R.,Want, F., Sockett, R.E.,Lambert, C., Hammond, M.C. "Production of 3', 3' -cGAMP by a Bdellovibrio bacteriovirus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility" PLoS Genetics (2022) 10, 1371.
- Tan, Z., Chan, C.H., Maleska, M., Banuelos Jara, B., Lohman, B.K., Ricks, N.J., Bond, D.R., Hammond, M.C. "The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens" Int. J. Mol. Sci. (2022) 23(3), 1183.
- Manna, S.*, Truong, J.*, Hammond, M.C. "Guanidine biosensors enable comparison of cellular turn-on kinetics of riboswitch-based biosensor and reporter" ACS Synth Biol (2021) 10, 3, 566-578 *co-first authors
- Manna, S., Kellenberger, C.A., Hallberg, Z.F., Hammond, M.C. "Live cell imaging using riboswitch-spinach tRNA fusions as metabolite-sensing fluorescent biosensors" Methods Mol Bio (2021) 2323, 121-140 (Invited Chapter).
- Kitto, R. Z.*, Dhillon, Y.*, Bevington, J.* et. al. Welch, C., McKay, C. P., Hammond, M. C. “Synthetic biological circuit tested in spaceflight” Life Sci Space Res (2021) 28, 57-65 *co-first authors
- Palmer, A. E., Hammond, M. C. “Editorial overview: Molecular imaging” Curr Opin Chem Biol (2020) 57, A5-A7.
- Kitto, R.Z., Christiansen, K.E., Hammond, M.C. "RNA-based fluorescent biosensors for live cell detection of bacterial sRNA" Biopolymers (2020) (Invited Paper) e23394.
- Yao, L., Fin A., Rovira, A.R., Su, Y., Dippel, A.B., Valderrama, J.A., Riestra, A.M., Nizet, V., Hammond, M.C., Tor, Y. "Tuning the innate immune response to cyclic dinucleotides using atomic mutagenesis" ChemBioChem (2020) 21, 18, 2595-2598.
- Anderson, W.A., Dippel, A.B., Maiden, M.M., Waters, C., Hammond, M. C. “Chemiluminescent sensors for quantitation of the bacterial second messenger cyclic di-GMP” Methods in Enzymology (2020) 640, 83-104 (Invited Chapter).
- Su, Y. and Hammond, M. C. “RNA-based fluorescent biosensors for live cell imaging of small molecules and RNAs” Curr Opin Biotech (2020) 63, 157-166 (Invited Review).
- Dippel, A. B.*, Anderson, W. A.*, Park, J. H., Yildiz, F. H., Hammond, M.C. "Development of ratiometric bioluminescent sensors for in vivo detection of bacterial signaling" ACS Chem Biol (2020) 15, 4, 904-914 *co-first authors
- Wright, T. A., Jiang, L., Park, J. J., Anderson, W. A., Chen, G., Hallberg, Z. F., Nan, B., and Hammond, M. C. "Second messengers and divergent HD-GYP enzymes regulate 3',3'-cGAMP signaling" Molecular Microbiology (2020) 113, 222-236.
- Wright, T. A., Dippel, A. B., Hammond, M. C. “Cyclic di-GMP signaling gone astray: cGAMP signaling via Hypr GGDEF and HD-GYP enzymes” In Chou, S.-H., Guilliani, N., Lee, V., Romling U. (ed), Microbial cyclic di-nucleotide signaling. (Invited Chapter).
- Dippel, A. B. and Hammond, M. C. "A poxin on both of your houses: Poxviruses degrade the immune signal cGAMP" Biochemistry (2019) 58, 19, 2387-2388 (Invited Viewpoint).
- Hallberg, Z. F.*, Chan, C. H.*, Wright, T. A., Park, J. J., Kranzusch, P. J., Doxzen,
K., Bond,
D. R., Hammond, M. C. “Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration” eLife (2019); e43959 *co-first authors - Villa, J.*, Su, Y.*, Contreras, L. M., Hammond, M. C. “Synthetic biology of small RNAs and riboswitches” (2019) 527-545. In Storz G, Papenfort K (ed), Regulating with RNA in Bacteria and Archaea. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.RWR-0007-2017 (Invited Book Chapter) *co-first authors
- Dippel, A. D., Anderson, W. A., Evans, R. S., Deutsch, S., Hammond, M. C. "Luminescent biosensors for detection of second messenger cyclic di-GMP" ACS Chem Bio (2018) 13, 1872-1879 (Invited Paper).
- Truong, J., Hsieh, Y.-F., Truong, L., Jia, G., Hammond, M.C. "Designing fluorescent biosensors using circular permutations of riboswitches", Methods (2018) 143, 102-109. (Invited Paper)
- Yeo, J., Dippel, A. B., Wang, X. C., Hammond, M. C. "In Vivo Biochemistry: Single-cell dynamics of cyclic di-GMP in E. coli in response to zinc overload" Biochemistry (2018) 57, 108-116.
- Yeo, J., Wang, X. C., Hammond, M. C. "Live flow cytometry analysis of c-di-GMP levels in single cell populations" Methods Mol Biol (2017) 1657, 111-130.
- Hallberg, Z. F., Su, Y., Kitto, R. Z., Hammond, M. C. "Engineering and in vivo applications of riboswitches" Annual Rev Biochem (2017) 86, 515-539.
- Bose, D.*, Su, Y.*, Marcus, A., Raulet, D. H., Hammond, M. C. "An RNA-based fluorescent biosensor for high-throughput analysis of the cGAS-cGAMP-STING pathway" Cell Chem Biol (2016) 23, 1539-1549. *co-first authors
- Wang, X. C., Wilson, S. C., Hammond, M. C. "Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP" Nucleic Acids Res (2016) 44, e139.
- Su, Y., Hickey, S. F., Keyser, S. G. L., Hammond, M. C. "In vitro and in vivo enzyme activity screening via RNA-based fluorescent biosensors for S-adenosyl-L-homocysteine (SAH)" J Am Chem Soc (2016) 138, 7040-7047.
- Hallberg, Z. F., Wang, X. C., Wright, T. A., Nan, B., Ad, O., Yeo, J., Hammond, M. C. "Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3', 3'-cGAMP)" Proc Natl Acad Sci (2016) 113, 1790-1795.
- Muller, R. Y., Hammond, M. C., Rio, D. C., Lee, Y. J. "An efficient method for electroporation of small interfering RNAs (siRNAs) into ENCODE Project Tier 1 GM12878 and K562 cell lines" J Biomol Tech (2015) 26, 142-149.
- Gonzalez, T. L., Liang, Y., Nguyen, B., Staskawicz, B. J., Loque, D., Hammond, M. C. "Tight regulation of plant immune responses by combining promoter and suicide exon elements" Nucleic Acids Res (2015) 43, 7152-7161.
- Kellenberger, C. A., Sales-Lee, J., Pan, Y., Gassaway, M. M., Herr, A. E., Hammond, M. C. "A minimalist biosensor: quantitation of cyclic di-GMP using the conformational change of a riboswitch aptamer" RNA Biol (2015) 12, 1189-1197.
- Kellenberger, C. A.*, Chen, C.*, Whiteley, A. T., Portnoy, D. A., Hammond, M. C. "RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP" J Am Chem Soc (2015) 137, 6432-6435.
- Ren, A., Wang, X. C., Kellenberger, C. A., Rajashankar, J. R., Jones, R., Hammond, M. C., Patel, D. J. "Structural basis for molecular discrimination by a 3', 3'-cGAMP sensing riboswitch" Cell Reports (2015), 11, 1-12.
- Kellenberger, C. A.*, Wilson, S. C.*, Hickey, S. F., Gonzalez, T. L., Su, Y., Hallberg, Z. F., Brewer, T. F., Iavarone, A. T., Carlson, H. K., Hsieh, Y. F., Hammond, M. C. "GEMM-I riboswitches from Geobacter sense the bacterial second messenger c-AMP-GMP" Proc Natl Acad Sci (2015) 112, 5383-5388. *co-first authors
- Kellenberger, C. A., Hallberg, Z. F., Hammond, M. C. "Live cell imaging using riboswitch-Spinach tRNA fusions as metabolite-sensing fluorescent biosensors" Methods in Mol Biol (2015) 1316, 87-103.
- Kellenberger, C. A., Hammond, M. C. "In vitro analysis of riboswitch-Spinach aptamer fusions as metabolite-sensing fluorescent biosensors" Methods Enz (2015) 550, 147-172.
- Pan, Y., Duncombe, T. A., Kellenberger, C. A., Hammond, M. C., Herr, A. E. "High-throughput electrophoretic mobility shift assays for quantitative analysis of molecular binding reactions" Analytical Chem (2014) 86, 10357-10364.
- Wilson, S. C., Cohen, D. T., Wang, X. C., Hammond, M. C. "A neutral pH thermal hydrolysis method for quantification of structured RNAs" RNA (2014) 20, 1153-1160.
- Hickey, S. F., Hammond, M. C. "Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs" Chem Biol (2014) 21, 345-356.
- Sadhu, M. J., Guan, Q., Li, F., Sales-Lee, J., Iavarone, A. T., Hammond, M. C., Cande, W. Z., Rine, J."Nutritional control of epigenetic processes in yeast and human cells" Genetics (2013) 195, 831-844.
- Diner, E. J., Burdette, D. L., Wilson, S. C., Monroe, K. M., Kellenberger, C. A., Hyodo, M., Hayakawa, Y., Hammond, M. C., Vance, R. E. "The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING" Cell Reports (2013), 3, 1355-1361.
- Leppek, K., Schott, J., Reitter, S., Poetz, F., Hammond, M. C., Stoecklin, G. "Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs" Cell (2013) 153, 869-881.
- Kellenberger, C. A., Wilson, S. C., Sales-Lee, J., Hammond, M. C. "RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP" J Am Chem Soc (2013) 135, 4906-4909.
- Karns, K., Vogan, J. M., Qin, Q., Hickey, S. F., Wilson, S. C., Hammond, M. C., Herr, A. E."Microfluidic screening of electrophoretic mobility shifts elucidates riboswitch binding function" J Am Chem Soc (2013), 135, 3136-3143.
- Hickey, S. F., Sridhar, M., Westermann, A. J., Qin, Q., Vijayendra, P., Liou, G., Hammond, M. C. "Transgene regulation in plants by alternative splicing of a suicide exon" Nucleic Acids Res (2012), 40, 4701-4710. Selected by NAR editors as a Featured Article
- Hammond, M. C. "A tale of two riboswitches" Nat Chem Biol (2011), 7, 342-3.
- Meyer, M. M., Hammond, M. C., Salinas, Y., Roth, A., Sudarsan, N., Breaker, R. R. "Challenges of ligand identification for riboswitch candidates" RNA Biol (2011), 8, 5-10.
- Block, K. F., Hammond, M. C., Breaker, R. R. "Evidence for widespread gene control function by the ydaO riboswitch candidate" J Bacter (2010), 192, 3983-9.
- Hammond, M. C., Wachter, A., Breaker, R. R. "A plant 5S rRNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs" Nat Struct and Mol Biol (2009), 16, 541-9.
- Weinberg, Z., Regulski, E. E., Hammond, M. C., Barrick, J. E., Yao, Z., Ruzzo, W. L., Breaker, R. R. "The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches" RNA (2008), 14, 822-8.
- Hammond, M. C., Bartlett, P. A. "Synthesis of amino acid-derived cyclic acyl amidines for use in beta-strand peptidomimetics" J Org Chem (2007), 72, 3104-07.
- Hammond, M. C., Harris, B. Z.; Lim, W. A., Bartlett, P. A. "Beta-strand peptidomimetics as potent PDZ ligands" Chem Biol (2006), 13, 1247-51.
- Sudarsan, N.*, Hammond, M. C.*, Block, K. F., Welz, R., Barrick, J. E., Roth, A., Breaker, R. R. "Tandem riboswitch architectures exhibit complex gene control functions" Science (2006), 314, 300-304. *co-first authors
