Kevin Hicks
Assistant Professor of Nutrition & Integrative Physiology and
Adjunct Assistant Professor of Biochemistry
Metabolism, Protein-metabolite Interactomics, Metabolic Signaling and Surveillance, Protein Biochemistry, Enzymology, Mass Spectrometry, Interactome Discovery Platforms

Molecular Biology Program
Education
B.S. University of Oregon
Ph.D. University of Washington
Research
Small molecule metabolites are conventionally viewed as the elementary constituents of life, functioning as cellular building blocks, energy currency, and waste products. Beyond anabolic and catabolic metabolism, it is increasingly evident that metabolites serve as signaling molecules, directly regulating protein function to provide a rapid and adaptive mechanism for metabolism to coordinate diverse cellular processes that are energetically or biosynthetically demanding. Furthermore, in healthy tissues, metabolism is internally balanced through evolved metabolic surveillance, more specifically, metabolite-mediated regulation of enzyme activity. Aberrant dysregulation of these regulatory protein-metabolite interactions likely contributes to the mechanisms driving many metabolically-dependent diseases including cancer, diabetes, heart disease, and inborn errors of metabolism. A deeper understanding of endogenous metabolite interactions with disease-relevant proteins will also benefit allosteric drug discovery. The true scale and function of the protein-metabolite interactome is almost completely unknown and represents a major void in our understanding of cellular biology and metabolism.
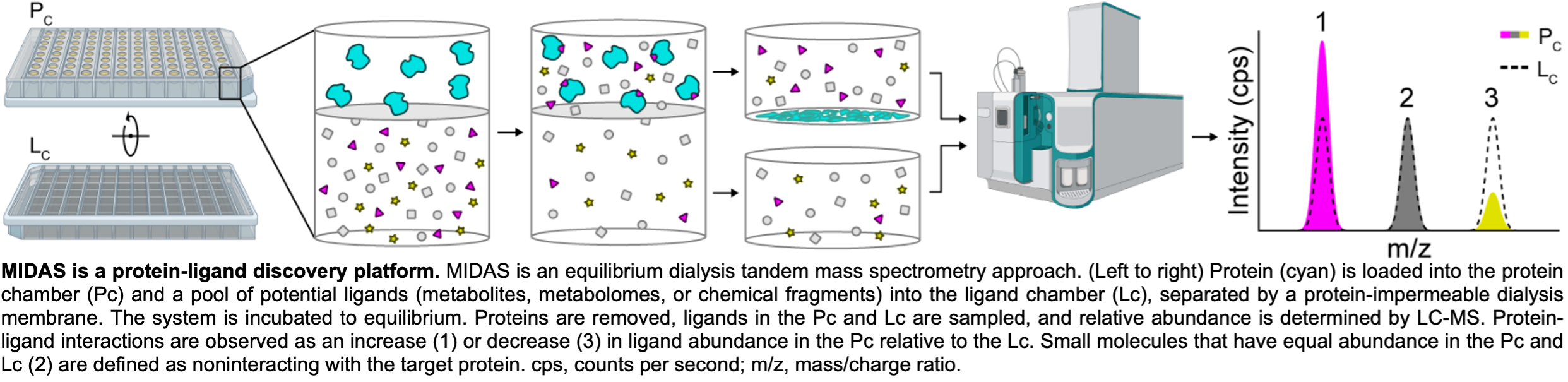
To begin to reveal the protein-metabolite interactome, we developed the MIDAS platform
to systematically identify interactions between proteins and metabolites (DOI: 10.1126/science.abm3452). The MIDAS platform is uniquely situated to identify low millimolar and stronger
binding between soluble proteins and metabolites from a multiplexed metabolite library
representative of the human metabolome. As such, the MIDAS platform is a powerful
tool to discover and investigate the form and function of the interactions spanning
the proteome and metabolome.
The Hicks lab is focused on two broad areas of research and development:
- Understanding how metabolism regulates metabolism. There are many well established examples of metabolites regulating specific metabolic
enzymes via allosteric and orthosteric interactions. These intra- and inter-pathway
protein-metabolite interactions serve as feedforward and feedback mechanisms to govern
metabolic pathway function, flux, and directionality. The extent of these regulatory
protein-metabolite interactions across metabolism is unknown. The Hicks lab uses innovative
and systematic mass spectrometry approaches to identify enzyme-metabolite interactions
and a compliment of molecular, biochemical, genetic, and cellular tools to reveal
their biological function. Systematically revealing the mechanisms of metabolic regulation
of human metabolic pathways is critical for a complete understanding of metabolism
and how diverse cellular processes are regulated or dysregulated in healthy and disease
states.
- Development of interactome discovery technologies. The strengths of the MIDAS platform can be harnessed to reveal interactions between molecules beyond soluble proteins and metabolites, opening new avenues of biological investigation. We are adapting the MIDAS platform to identify membrane protein-metabolite, protein-metabolome, protein-peptide, protein-chemical fragment, and RNA-metabolite interactions using targeted and non-targeted liquid chromatography mass spectrometry. These interactomics platforms will broaden our understanding of the scope, depth, and functions of interacting biological networks, provide inclusive discovery platforms for the scientific community, and catalyze therapeutic discovery.
References (Selected Publications)
- Hicks KG, Cluntun AA, Schubert HL, Hackett SR, Berg JA, Leonard PG, Ajalla Aleixo MA, Zhou Y, Bott AJ, Salvatore SR, Chang F, Blevins A, Barta P, Tilley S, Leifer A, Guzman A, Arok A, Fogarty S, Winter JM, Ahn HC, Allen KN, Block S, Cardoso IA, Ding J, Dreveny I, Gasper WC, Ho Q, Matsuura A, Palladino MJ, Prajapati S, Sun P, Tittmann K, Tolan DR, Unterlass J, VanDemark AP, Vander Heiden MG, Webb BA, Yun CH, Zhao P, Wang B, Schopfer FJ, Hill CP, Nonato MC, Muller FL, Cox JE, Rutter J. Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase. Science. 2023 Mar 10;379(6636):996-1003. doi: 10.1126/science.abm3452. Epub 2023 Mar 9. PubMed PMID: 36893255; PubMed Central PMCID: PMC10262665.
- Nielson JR, Nath AK, Doane KP, Shi X, Lee J, Tippetts EG, Saha K, Morningstar J, Hicks KG, Chan A, Zhao Y, Kelly A, Hendry-Hofer TB, Witeof A, Sips PY, Mahon S, Bebarta VS, Davisson VJ, Boss GR, Rutter J, MacRae CA, Brenner M, Gerszten RE, Peterson RT. Glyoxylate protects against cyanide toxicity through metabolic modulation. Sci Rep. 2022 Mar 23;12(1):4982. doi: 10.1038/s41598-022-08803-y. PubMed PMID: 35322094; PubMed Central PMCID: PMC8943054.
- Hao Q, Heo JM, Nocek BP, Hicks KG, Stoll VS, Remarcik C, Hackett S, LeBon L, Jain R, Eaton D, Rutter J, Wong YL, Sidrauski C. Sugar phosphate activation of the stress sensor eIF2B. Nat Commun. 2021 Jun 8;12(1):3440. doi: 10.1038/s41467-021-23836-z. PubMed PMID: 34103529; PubMed Central PMCID: PMC8187479.
- Bezerra GA, Holenstein A, Foster WR, Xie B, Hicks KG, Bürer C, Lutz S, Mukherjee A, Sarkar D, Bhattacharya D, Rutter J, Talukdar A, Brown PJ, Luo M, Shi L, Froese DS, Yue WW. Identification of small molecule allosteric modulators of 5,10-methylenetetrahydrofolate reductase (MTHFR) by targeting its unique regulatory domain. Biochimie. 2021 Apr;183:100-107. doi: 10.1016/j.biochi.2021.01.007. Epub 2021 Jan 18. PubMed PMID: 33476699; PubMed Central PMCID: PMC8040968.
- Bezerra GA, Foster WR, Bailey HJ, Hicks KG, Sauer SW, Dimitrov B, McCorvie TJ, Okun JG, Rutter J, Kölker S, Yue WW. Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism. IUCrJ. 2020 Jul 1;7(Pt 4):693-706. doi: 10.1107/S205225252000696X. eCollection 2020 Jul 1. PubMed PMID: 32695416; PubMed Central PMCID: PMC7340257.
- Hicks KG, Delbecq SP, Sancho-Vaello E, Blanc MP, Dove KK, Prost LR, Daley ME, Zeth K, Klevit RE, Miller SI. Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence. Elife. 2015 May 23;4:e06792. doi: 10.7554/eLife.06792. PubMed PMID: 26002083; PubMed Central PMCID: PMC4473727.
- Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012 Jul 3;20(7):1177-88. doi: 10.1016/j.str.2012.04.021. Epub 2012 Jun 14. PubMed PMID: 22705207; PubMed Central PMCID: PMC3392440.
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011 Dec;82(5):1277-90. doi: 10.1111/j.1365-2958.2011.07889.x. Epub 2011 Nov 4. PubMed PMID: 22017253; PubMed Central PMCID: PMC3590308.
- Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology (Reading). 2011 Sep;157(Pt 9):2445-2455. doi: 10.1099/mic.0.049353-0. Epub 2011 May 20. PubMed PMID: 21602215; PubMed Central PMCID: PMC3352171.
- Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010 Jan 21;7(1):25-37. doi: 10.1016/j.chom.2009.12.007. PubMed PMID: 20114026; PubMed Central PMCID: PMC2831478.
