Martin Horvath
Associate Professor of Biological Sciences
Structural Biology, Biochemistry, DNA repair, Eye Health

Molecular Biology Program
Biological Chemistry Program
Education
B.S. Brown University
Ph.D. University of Chicago
Research
Structural Biology & Biochemistry. My research explores how proteins work, and how molecular interactions establish emergent properties of specificity, allostery, and catalysis. With x-ray crystallography, I obtained structures for DNA enzymes with DNA, including the first structure of a single-stranded DNA-protein complex found at the telomere. With Cryo-EM I aim to build structures for membrane proteins. My expertise helps my colleagues think about biological systems in terms of molecular mechanisms.
References
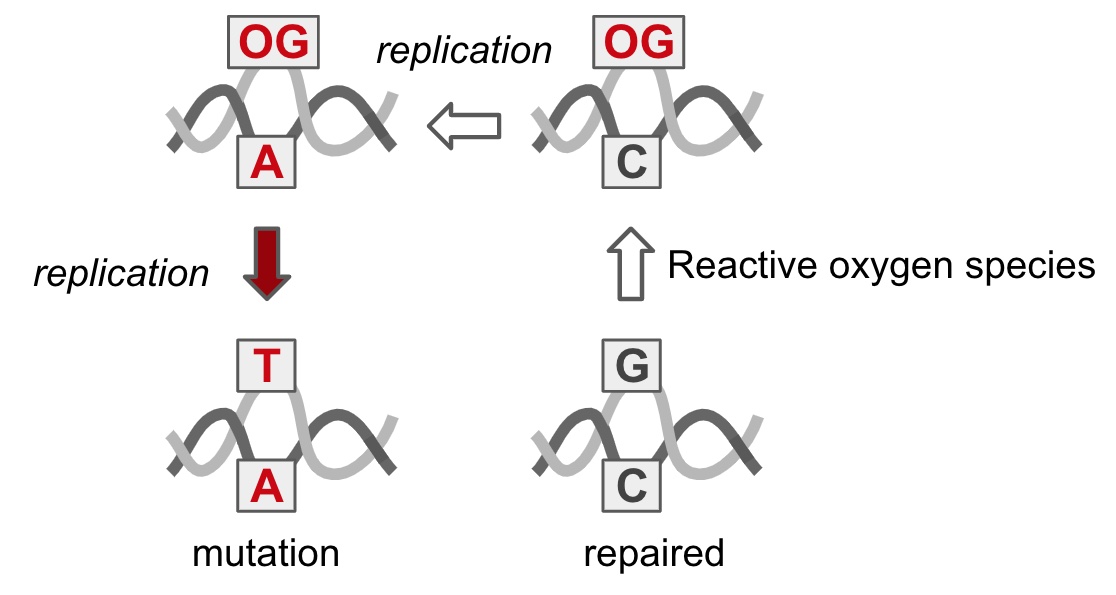
- Utzman, P. H.; Mays, V. P.; Miller, B. C.; Fairbanks, M. C.; Brazelton, W. J.; Horvath, M. P. Metagenome Mining and Functional Analysis Reveal Oxidized Guanine DNA Repair at the Lost City Hydrothermal Field. bioRxiv April 9, 2023, p 2023.04.05.535768. https://doi.org/10.1101/2023.04.05.535768.
- Utzman, P. H.; Mays, V.; Miller, B.; Brazelton, W. J.; Horvath, M. P. Metagenome Mining and Functional Analysis Reveal Oxidative DNA Repair at the Lost City Hydrothermal Field. PLoS ONE (in revision) 2023.
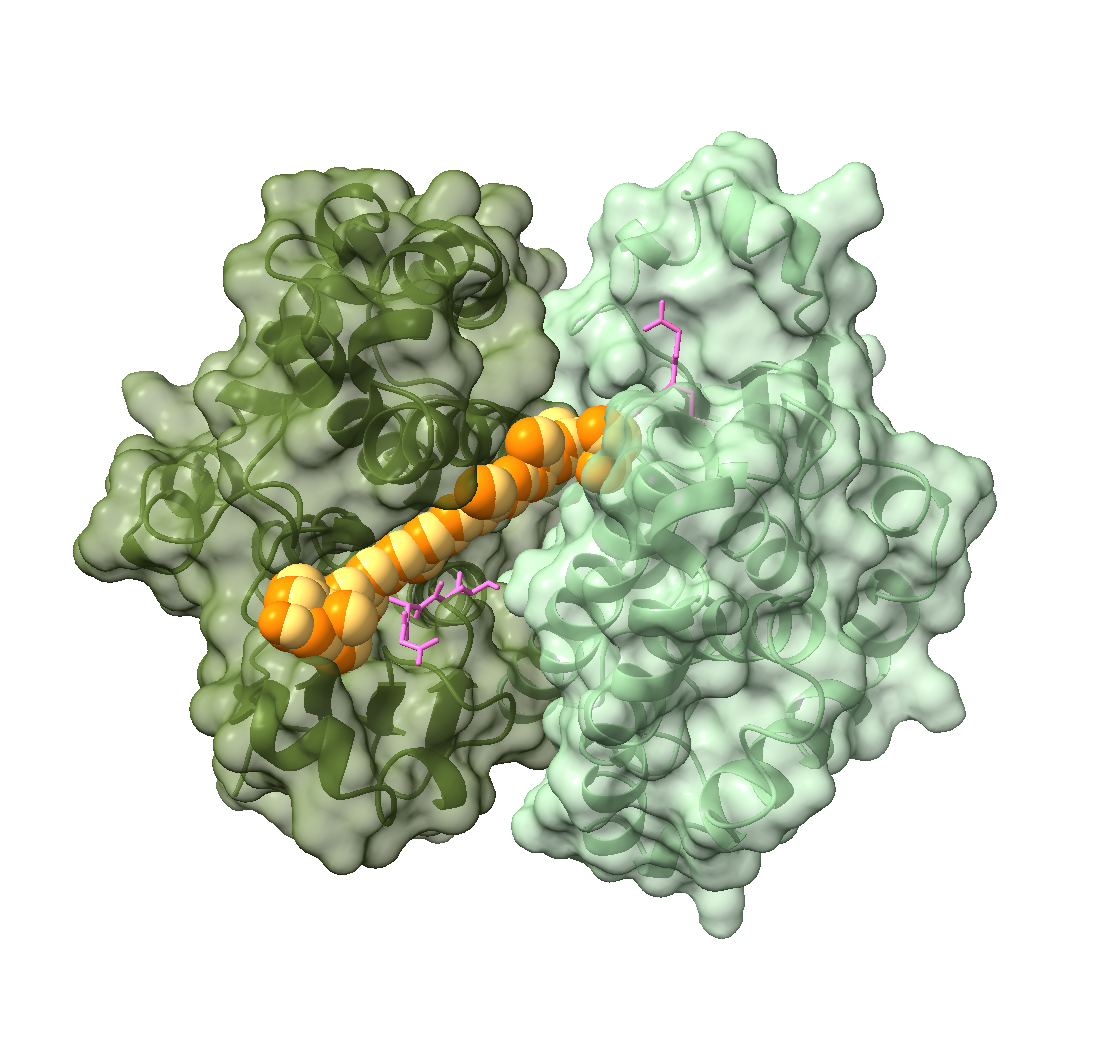
- Demir, M.; Russelburg, L. P.; Lin, W.-J.; Trasviña-Arenas, C. H.; Huang, B.; Yuen, P. K.; Horvath, M. P.; David, S. S. Structural Snapshots of Base Excision by the Cancer-Associated Variant MutY N146S Reveal a Retaining Mechanism. Nucleic Acids Res. 2023, gkac1246. https://doi.org/10.1093/nar/gkac1246.
- Russelburg, L. P.; O’Shea Murray, V. L.; Demir, M.; Knutsen, K. R.; Sehgal, S. L.; Cao, S.; David, S. S.; Horvath, M. P. Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY. ACS Chem. Biol. 2020, 15 (1), 93–102. https://doi.org/10.1021/acschembio.9b00639.
- Woods, R. D.; O’Shea, V. L.; Chu, A.; Cao, S.; Richards, J. L.; Horvath, M. P.; David, S. S. Structure and Stereochemistry of the Base Excision Repair Glycosylase MutY Reveal a Mechanism Similar to Retaining Glycosidases. Nucleic Acids Res. 2016, 44 (2), 801–810. https://doi.org/10.1093/nar/gkv1469.
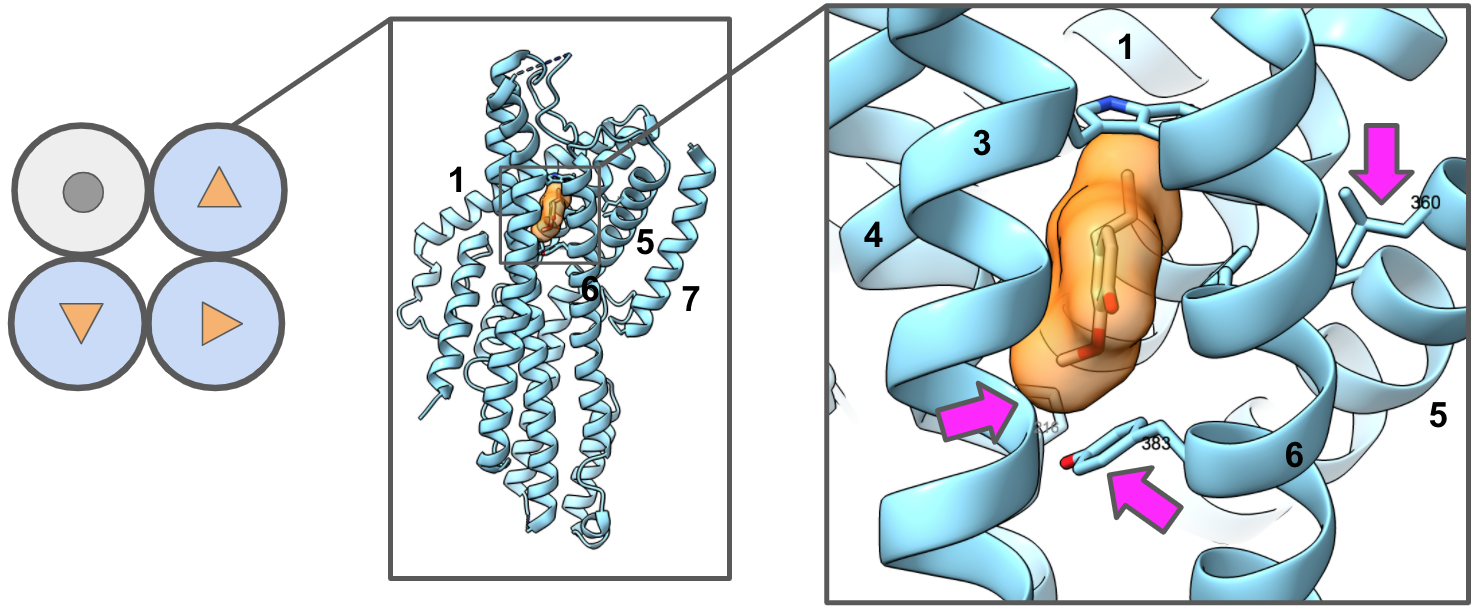
- Shyam, R.; Gorusupudi, A.; Nelson, K.; Horvath, M. P.; Bernstein, P. S. RPE65 Has an Additional Function as the Lutein to Meso-Zeaxanthin Isomerase in the Vertebrate Eye. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (41), 10882–10887. https://doi.org/10.1073/pnas.1706332114.
- Horvath, M. P.; George, E. W.; Tran, Q. T.; Baumgardner, K.; Zharov, G.; Lee, S.; Sharifzadeh, H.; Shihab, S.; Mattinson, T.; Li, B.; Bernstein, P. S. Structure of the Lutein-Binding Domain of Human StARD3 at 1.74 Å Resolution and Model of a Complex with Lutein. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72 (Pt 8), 609–618. https://doi.org/10.1107/S2053230X16010694.
- Curtice, K. J.; Leavitt, L. S.; Chase, K.; Raghuraman, S.; Horvath, M. P.; Olivera, B. M.; Teichert, R. W. Classifying Neuronal Subclasses of the Cerebellum through Constellation Pharmacology. J. Neurophysiol. 2016, 115 (2), 1031–1042. https://doi.org/10.1152/jn.00894.2015.
- Dy, C. Y.; Buczek, P.; Imperial, J. S.; Bulaj, G.; Horvath, M. P. Structure of Conkunitzin-S1, a Neurotoxin and Kunitz-Fold Disulfide Variant from Cone Snail. Acta Crystallogr. D Biol. Crystallogr. 2006, 62 (Pt 9), 980–990. https://doi.org/10.1107/S0907444906021123.
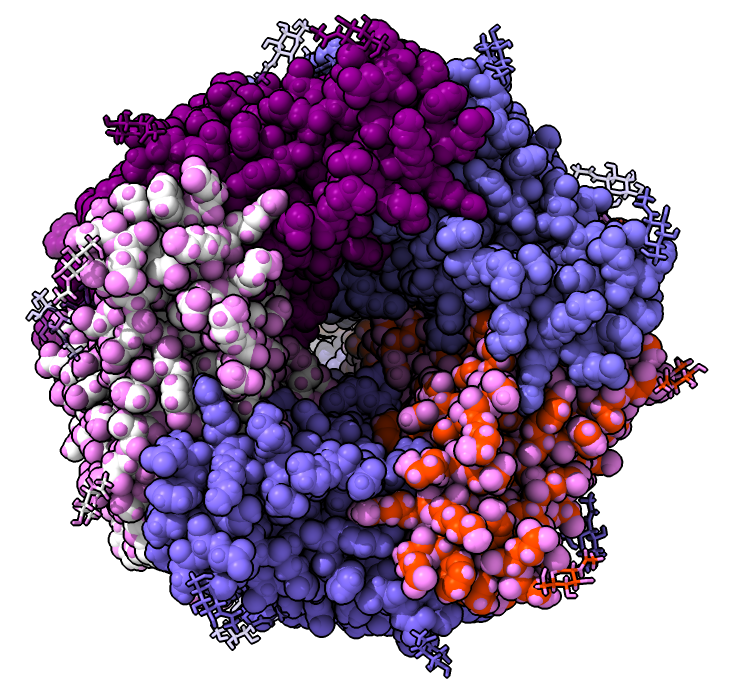
- Buczek, P.; Horvath, M. P. Structural Reorganization and the Cooperative Binding of Single-Stranded Telomere DNA in Sterkiella Nova. J. Biol. Chem. 2006, 281 (52), 40124–40134. https://doi.org/10.1074/jbc.M607749200.
- Horvath, M. P.; Schweiker, V. L.; Bevilacqua, J. M.; Ruggles, J. A.; Schultz, S. C. Crystal Structure of the Oxytricha Nova Telomere End Binding Protein Complexed with Single Strand DNA. Cell 1998, 95 (7), 963–974. https://doi.org/10.1016/s0092-8674(00)81720-1.