Mark Metzstein
Associate Professor of Human Genetics
Drosophila Genetics

Molecular Biology Program
Education
B.A. University of Cambridge
Ph.D. Massachusetts Institute of Technology
Research
In my laboratory, we are interested in two biological processes: cell morphogenesis and regulation of RNA stability. To analyze these, we use Drosophila genetics and molecular biology, along with microscopy, cell biological and genomic approaches.
Drosophila Tracheal Cell Morphogenesis
We are interested in the mechanisms by which cells form their shapes to build tissues and organs. To address this question, we are studying the development of the Drosophila tracheal system, an interconnected network of epithelial tubes through which oxygen is delivered to internal tissues. The branching and tubulogenesis processes in one tracheal cell type, terminal cells, are of particular interest. These cells show considerable morphological complexity: each cell forms dozens of fine subcellular branches by a process that involves cytoplasmic extension and bifurcation, and each branch forms an intracellular lumen through which air flows. This complexity at the single cell level, combined with the genetic and molecular tools available in Drosophila, represent an excellent opportunity to ask and answer questions about cellular morphogenesis.
To address how the tracheal terminal cells form, we have previously carried out large scale genetic screens for mutants affecting terminal cell development. We are now focusing on the molecular and cell biological functions of the genes defined by this screen. For example, analysis of one of our mutants (out of gas) has suggested a novel role for a multivesicular body-like compartment in intracellular lumen formation. Another gene, Zpr1, suggests a link between lumen formation and EGF signaling. Finally, from our mutants we have identified a dual role for PAR-complex genes (par-6, aPKC, and bazooka) in lumen formation and branching.
To complement our genetic studies, we are carrying out a detailed ultrastructural studies of tracheal terminal cells. We are developing novel electron microscope techniques to preserve membrane structure within terminal cells and to identify mutant cells in genetically mosaic animals. Using these approaches, we will be able to better understand the membrane dynamics the occur to make the lumen, and be able characterize our mutants at the ultrastructural level.
Genetic and Genomic Analysis of Nonsense Mediated mRNA Decay
The NMD pathway monitors mRNAs to identify transcripts that contain premature stop codons and to target these RNAs for rapid destruction. We are using Drosophila genetics to identify the cis and trans acting factors that are used to identify such aberrant mRNAs. We are carrying out large scale genetic screens to identify mutations affecting the NMD process and have now identified numerous such mutations. Most of these are in genes known in other organisms to be required for NMD, while some are likely to define novel components of the pathway. We are now carrying out genetic and molecular analysis of our identified alleles and extending our screens to cover the whole Drosophila genome.
We are also performing a transcriptome analysis of NMD mutant animals to identify the repertoire of endogenous target genes. Bioinformatic characterization of these targets should reveal the set of cis-acting signals which direct mRNAs for degradation via the NMD pathway. Additionally, the targets should reveal the tissue and cellular requirements for NMD in Drosophila development and physiology.
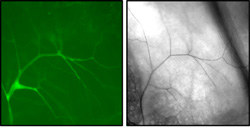
Morphology of a single Drosophila tracheal terminal cell. Left: cytoplasmically localized GFP expressed in the terminal cell shows numerous subcellular branches generated by the cell. Right: brightfield microscopy of the same cell reveals an air-filled luminal tube running within each branch.
References
- Nelson JO, Moore KA, Chapin A, Hollien J, Metzstein MM (2016) Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife. Mar 8:5. pii: e12876
- Nikolova LS, Metzstein MM (2015) Intracellular lumen formation in Drosophila proceeds via a novel subcellular compartment. Development 142(22):3964-73
- Jones TA, Nikolova LS, Schjelderup A, Metzstein MM (2014) Exocyst-mediated membrane trafficking is required for branch outgrowth in Drosophila tracheal terminal cells. Dev Biol 390(1):41-50
- Chapin A, Hu H, Rynearson SG, Hollien J, Yandell M, Metzstein MM (2014) In vivo determination of direct targets of the nonsense-mediated decay pathway in Drosophila. G3 (Bethesda) 4(3):485-96
- Jones TA, Metzstein MM (2013) Examination of Drosophila larval tracheal terminal cells by light microscopy. J Vis Exp (77):e50496
- Ruiz OE, Nikolova LS, Metzstein MM (2012) Drosophila Zpr1 (Zinc finger protein 1) is required downstream of both EGFR and FGFR signaling in tracheal subcellular lumen formation. PLoS ONE 7(9):e45649
- Frizzell KA, Rynearson SG, Metzstein MM (2012) Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6. RNA 18(8):1475-1486
- Jones TA, Metzstein MM (2011) A Novel Function for the PAR Complex in Subcellular Morphogenesis of Tracheal Terminal Cells in Drosophila melanogaster. Genetics 189(1):153-164
- Chan AC, Drakos SG, Ruiz OE, Smith AC, Gibson CC, Ling J, Passi SF, Stratman AN, Sacharidou A, Revelo MP, Grossmann AH, Diakos NA, Davis GE, Metzstein MM,Whitehead KJ, Li DY (2011) Mutations in 2 distinct genetic pathways result in cerebral cavernous malformations in mice. J Clin Invest 121(5):1871-1881
- Long AA, Mahapatra C, Woodruff III EA, Rohrbough J, Leung H-T, Shino S, An L, Doerge RW, Metzstein MM, Pak WL, Broadie K (2010) The nonsense-mediated decay pathway is critical for maintaining synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci 123(19):3303-3315
- Nien C-Y, Liang H-L, Liu H-Y, Metzstein MM, Kirov N, Rushlow C (2008) The zinc finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456(7220):400-403
- Metzstein MM, Krasnow M (2006) Functions of the Nonsense-Mediated RNA Decay Pathway in Drosophila Development. PLoS Genet 2(12):180
- Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA (2003) Branching morphogenesis of the Drosophila tracheal system. [Review]. Annu Rev Cell Dev Biol 19:623-647
