Peter Shen
Associate Professor of Biochemistry
Protein Homeostasis, Protein Structure, cryo-EM

Molecular Biology Program
Biological Chemistry Program
Education
B.S. Brigham Young University
Ph.D. Brigham Young University
Research
Protein Quality Control
Protein homeostasis is central to maintaining a healthy cellular environment. All proteins rely on quality control mechanisms to ensure proper synthesis, folding, function, and turnover. Dysregulation along these pathways contribute to some of the most devastating diseases in humans, including cancer and neurodegeneration. My lab seeks to resolve the molecular mechanisms that are responsible for maintaining proper protein quality control. The mechanisms that we uncover will inform structure-guided approaches to the development of therapeutics that counteract quality control failure.
Our active areas of focus include protein synthesis (ribosomes), folding (chaperonins), and unfolding (AAA+ ATPases). A common theme among these projects is their involvement of large and dynamic macromolecular complexes. We specialize in isolating and characterizing such complexes directly from their native sources and in their active states. We also apply these methods to study how these processes break down in the context of disease, such as from a result of mutations and viral infection.
Seeing is Believing
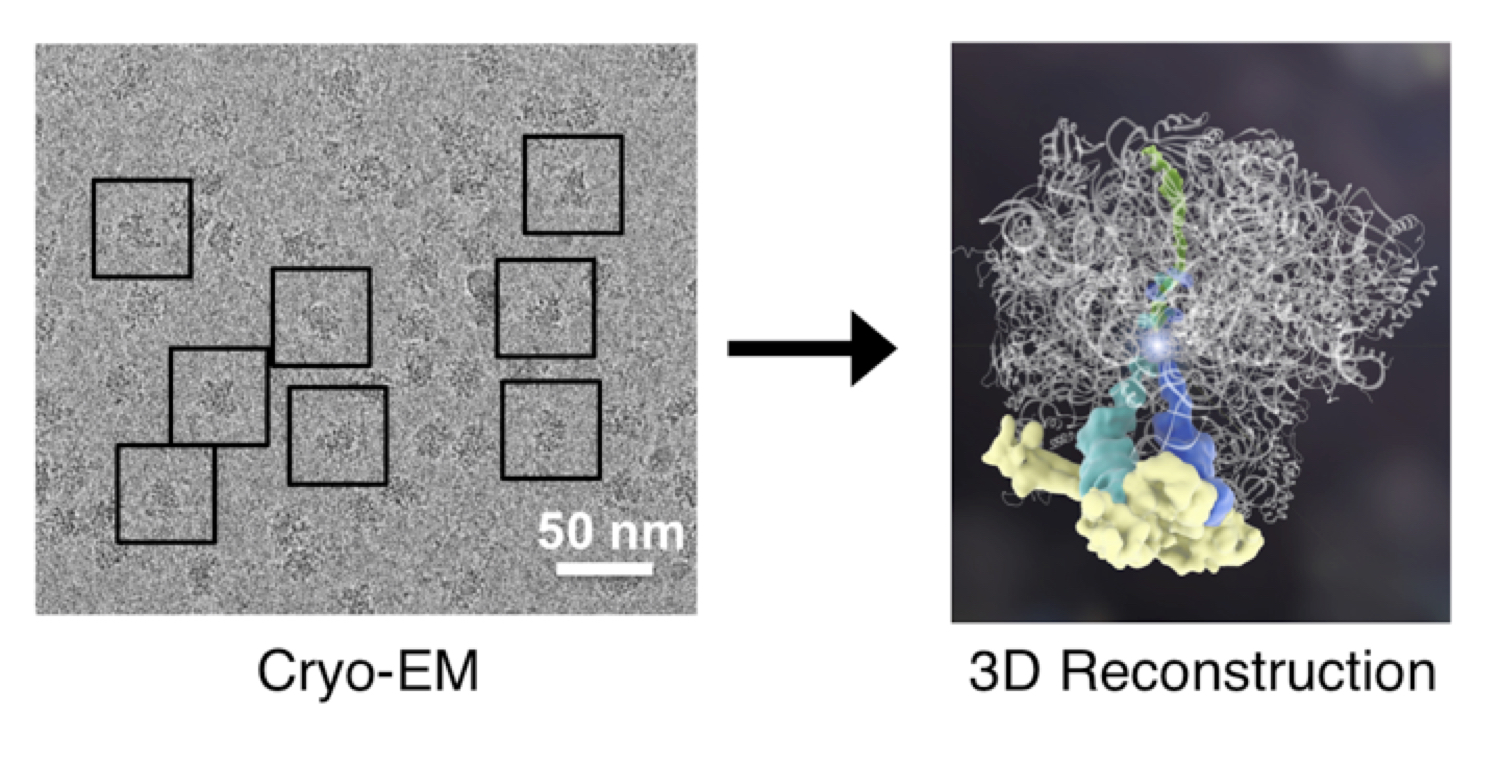
All cellular processes depend on atomic-scale interactions. In order to understand molecular mechanism, my lab specializes in cryo-EM to resolve high-resolution 3D structures. Cryo-EM is uniquely suited for working with structurally heterogeneous samples because a single sample often reveals a range of structural states that can then be pieced together to deduce their mechanisms. The ability to directly visualize these biological molecules will enable us to understand how multiple components come together to carry out their evolutionarily tuned functions. Learn more about cryo-EM at https://CryoEM101.org.
References
- Wang, S, Sass MI, Kwon Y, Ludlam WG, Smith TM, Carter EJ, Gladden NE, Riggi M, Iwasa JH, Willardson BM*, Shen PS*. Visualizing the chaperone-mediated folding trajectory of the G protein β5.Biorxiv (2023) doi:10.1101/2023.05.04.539424. [Preprint]. Posted May 4, 2023. (*co-corresponding author)
- Xu Y, Han H, Cooney I, Guo Y, Moran NG, Zuniga NR, Price JC, Hill CP*, Shen PS*. Active conformation of the p97-p47 unfoldase complex. Nat. Commun. 2022; 2640:https://doi.org/10.1038/s41467-022-30318-3 (*co-corresponding author)
- Rollins MG, Shasmal M, Meade N, Astar, H, Shen PS*, Walsh D*. Negative charge in the RACK1 loop broadens the translational capacity of the human ribosome. Cell Rep. 2021 Sep 7;36(10):109663. (*co-corresponding author)
- Cooney I, Han H, Stewart MG, Carson RH, Hansen DT, Iwasa JH, Price JC, Hill CP*, Shen PS*. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019 Jun 27 (*co-corresponding author)
- Han H, Fulcher JM, Dandey VP, Iwasa JH, Sundquist WI, Kay MS, Shen PS*, Hill CP*. Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases. Elife. 2019 Jun 11;8 (*co-corresponding author)
- Sinha NK, Iwasa J, Shen PS*, Bass BL*. Dicer uses distinct modules for recognizing dsRNA termini.Science. 2018 Jan 19:359(6373):329–34. (*co-corresponding author)
- Han H, Monroe N, Sundquist WI*, Shen PS*, Hill CP*. The AAA ATPase Vps4 Binds ESCRT-III Substrates Through a Repeating Array of Dipeptide-binding Pockets.Elife. 2017 pii: e31324. (*co-corresponding author)
- Shen PS*, Yang X*, DeCaen PG, Liu X, Bulkley D, Clapham DE, Cao E. The Structure of Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 2016 Oct 20:167(3):763–73.
- Shen PS, Park J, Qin Y, Parsawar K, Li X, Larson M, Cox J, Cheng Y, Lambowitz AM, Weissman JS, Brandman O, Frost A. Rqc2p and the 60S ribosome mediate mRNA-independent elongation of nascent chains. Science. 2015 Jan 2;347(6217):75–8.
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012 Nov 21;151(5):1042–54.