Xiaoxu Yang
Assistant Professor of Human Genetics
Mosaicism, Genomics, Computational biology, Variant calling, Bioinformatics, Human genetics, Development, Genome mutation

Molecular Biology Program
Education
B.S. Beijing Normal University, Beijing, China
Ph.D. Peking University, Beijing, China
Research
Understand human disease and development with genomic mosaicism
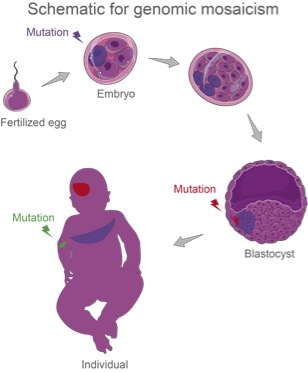
From a fertilized egg, all our cells acquire mutations at all times, till the end of life. Most of the mutations are repaired by our body, but some remain in our genome and get passed from cell to cell during embryonic development, upon tissue self-renewal, or after environmental exposure. Genome mosaicism reflects the phenomenon that cells from the same zygote have different genomic sequences.
Unlike other types of mutations that appear in all cells of the body, mosaic mutations are difficult to detect due to the low presentations in samples we can obtain. High accuracy and high sensitivity low-fraction detection methods, both experimental and computational ones, are in high demand for this field.
Patterns of genomic mosaicism, as a combinatory consequence of various mutation procedures as described above, could help us understand the biological processes. For example, mosaic mutations shared between different tissues could serve as markers to trace when two different cell populations shared a common progenitor, which largely remained unsolved in human development.
Mosaicism has been reported to directly contribute to hundreds of documented human disorders. Many disorders related to genomic mosaicism have an obvious mosaic nature, such as focal epilepsies, patchy skin, or bone disorders that only affect some, but not all of the cells. There are, however, disorders caused by mosaic mutations that are not so obvious to suspect.
Mosaic mutations can also be transmitted to the next generation, by definition, only part of the cells carrying the mutation might lead to severe consequences for the next generation. Thus understanding genomic mosaicism between generations will help us understand the genomic health of ourselves and our children.
Methodolodies to study mosaicism:

With the fast accumulation of human genomics and genetics data, the demand for method development for mosaic mutations has emerged unprecedently. Accurate detection and quantification of mosaic variants lay the foundation of mosaicism-related research. We pioneered the study of genomic mosaicism, I established computational methods for the identification of postzygotic mutation including DeepMosaic (Nat Biotech 2023), and MFMS (Cell 2021). I also developed experimental pipelines PASM/TASeq (Hum Mutat 2015), MPAS/single cell MPAS (Nature 2022, 2024), and mDDPCR (Sci Rep 2017) for the quantification of mosaic mutations in clinical and base research studies. We are also seeking to develop highly sensitive experimental variant quantification and validation strategies.
Germline mosaicism for transmisison risk
Detectable mosaic variants in the parental germline might be inherited by the offspring and cause a severe impact on the children's health. One of the major research interests of the Yang Lab is how parental germline mosaicism, especially sperm mosaic mutations, is affecting the health of the next generation. On the other hand, male-driven evolution is a fundamental genetic driving force of evolution and speciation, understanding the impact of genomic stability between generations will help us to understand how we are humans, and where are we going. We are employing state-of-the-art sequencing and data mining technologies to profile mosaic mutations from sperm in order to understand how environments, aging, and lifestyles are affecting the human sperm genome and shaping the mutational landscape.With the methods we developed, I pioneered the field to estimate the genetic risk of severe neurological disorders from the parental germline. I found more than 10% of seemingly de novo disease-causing mutations were actually inherited from paternal sperm mosaicism (Hum Mutat 2015, J Med Genet 2018, Clin Genet 2019, Genet Med 2019).weI predicted one in 15 normal males harbor deleterious clonal sperm mosaicism which will be transmitted to offspring and create a life-long genetic risk (Cell 2021, eLife 2022).
Somatic mosaicism and lineage reconstruction in Humans
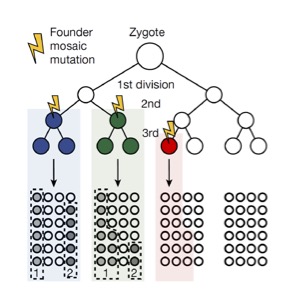
Lineage markers with dye, genetic barcodes, or isotope pulse-chase have been used to study the development of model organisms for decades. These interventions, however, are not suitable for human development. Through embryonic development, body growth, and tissue renewal, somatic mutations accumulate across the genome, if mutations fail to be repaired, they will be inherited by daughter cells and serve as neutral lineage markers for development. By using these 'molecular paints' in our body, we consider mosaic variant as a unique barcode to reconstruct human development and find important developmental clonal distributions that might be unique to humans.
We proposed the idea that mosaic variants are either shared or unique to tissues within the same individual (PLoS Genet 2018). Based on this, we further established the idea of using mosaic mutations as a neutral recorder to study human embryonic development. I opened this new direction to use mosaic mutations to study human neocortical development and found that left-right separation between cortical hemispheres occurred before anterior-posterior lobar separations in humans (Nature 2022) and further convincing showed with mosaic variant barcoding analysis dorsally derived inhibitory neuron in human cortex (Nature 2024).
Mosaicism in human disease and health

Somatic mosaic variants are the intrinsic cause of most cancers and more than 200 non-cancer disorders. Growing evidence has shown that mosaic mutations are correlated with neurological, metabolic, and age-related disorders. Understanding the mosaic mutational pathogenicity of disease is the key to early diagnosis and treatment.
With these understandings about cortical development, I detected mTOR and non-mTOR mosaic mutations causing malformation of cortical development (Nat Genet 2023). Based on my rich experience of mosaicism in non-cancer diseases and normal development, I established the genetic knowledge base MosaicBase to facilitate biomedical research in mosaicism study (GPB 2020).
References
Selected Publications
- Muhammad Zubair, Xiaoxu Yang (2025). Dipping Into the Phenotypic Implications of Mosaic Variants. [Editorial]. Neurology Genetics, 11(2), e200256.
- Camila Araújo Bernardino Garcia*, Muhammad Zubair*, Marcelo Volpon Santos, Sang Hyun Lee, Alfred Graham, Valentina Stanley, Renee D. George, Joseph G. Gleeson, Hélio Rubens Machado#, Xiaoxu Yang# (2025). Identification of Novel Mosaic Variants in Focal Epilepsy-Associated Patients’ Brain Lesions. Genes. 16 (4), 421.
- Vong KI*, Alvarez YD, Noel G, Barton ST, Chung C, Howarth R, Meave N, Zhang Q, Jiwani F, Barrows C, Patel A, Wang JX, Chi N, Kingsmore SF, White MD, Yang X*,#, Gleeson JG# (2024). Genomic mosaicism reveals developmental organization of trunk neural crest-derived ganglia. bioRxiv. https://doi.org/10.1101/2024.09.25.615004
- Chung C*, Yang X*, Hevner RF, Kennedy K, Vong KI, Liu Y, Patel A, Nedunuri R, Barton ST, Barrows C, Stanley V, Mittal S, Breuss MW, Schlachetzki JCM, Gleeson JG (2024). Cell-type-resolved mosaicism reveals clonal dynamics of the human forebrain. Nature. 629 (8011), 384-392
- Yang X*, Xu X*, Breuss MW, Antaki D, Ball LL, Chung C, Shen J, Li C, George RD, Wang Y, Bae T, Cheng Y, Abyzov A, Wei L, Alexandrov LB, Sebat JL, NIMH Brain Somatic Mosaicism Network, Gleeson JG (2023). Control-independent mosaic single nucleotide variant detection with DeepMosaic. Nat Biotechnol, 41(6), 870-877.
- Chung C*, Yang X*, Bae T, Vong KI, Mittal S, Donkels C, Westley Phillips H, Li Z, Marsh APL, Breuss MW, Ball LL, Garcia CAB, George RD, Gu J, Xu M, Barrows C, James KN, Stanley V, Nidhiry AS, Khoury S, Howe G, Riley E, Xu X, Copeland B, Wang Y, Kim SH, Kang HC, Schulze-Bonhage A, Haas CA, Urbach H, Prinz M, Limbrick DD Jr, Gurnett CA, Smyth MD, Sattar S, Nespeca M, Gonda DD, Imai K, Takahashi Y, Chen HH, Tsai JW, Conti V, Guerrini R, Devinsky O, Silva WA Jr, Machado HR, Mathern GW, Abyzov A, Baldassari S, Baulac S, Focal Cortical Dysplasia Neurogenetics Consortium, Brain Somatic Mosaicism Network, Gleeson JG (2023). Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development. Nat Genet, 55(2), 209-220. (Cover article)
- Breuss MW*,#, Yang X*, Stanley V, McEvoy-Venneri J, Xu X, Morales AJ, Gleeson JG (2022). Unbiased mosaic variant assessment in sperm: a cohort study to test predictability of Elife, 11 e78459.
- Breuss MW*, Yang X*, Schlachetzki JCM*, Antaki D*, Lana AJ, Xu X, Chung C, Chai G, Stanley V, Song Q, Newmeyer TF, Nguyen A, O'Brien S, Hoeksema MA, Cao B, Nott A, McEvoy-Venneri J, Pasillas MP, Barton ST, Copeland BR, Nahas S, Van Der Kraan L, Ding Y, NIMH Brain Somatic Mosaicism Network, Glass CK, Gleeson JG (2022). Somatic mosaicism reveals clonal distributions of neocortical development. Nature, 604(7907), 689-696.
- Yang X*, Breuss MW*, Xu X, Antaki D, James KN, Stanley V, Ball LL, George RD, Wirth SA, Cao B, Nguyen A, McEvoy-Venneri J, Chai G, Nahas S, Van Der Kraan L, Ding Y, Sebat J, Gleeson JG (2021). Developmental and temporal characteristics of clonal sperm mosaicism. Cell, 184(18), 4772-4783.e15.
- Breuss MW*, Yang X*, Gleeson JG (2021). Sperm mosaicism: implications for genomic diversity and disease. [Review]. Trends Genet, 37, (10), 890-902.
- Yang X*, Yang C*, Zheng X*, Xiong L, Tao Y, Wang M, Ye AY, Wu Q, Dou Y, Luo J, Wei L#, Huang AY# (2020). MosaicBase: A Knowledgebase of Postzygotic Mosaic Variants in Noncancer Disease-related and Healthy Human Genomics Proteomics Bioinformatics, 18(2), 140-149.
- Yang X*, Yang X*, Chen J, Li S, Zeng Q, Huang AY, Ye AY, Yu Z, Wang S, Jiang Y, Wu X, Wu Q, Wei L#, Zhang Y# (2019). ATP1A3 mosaicism in families with alternating hemiplegia of childhood. Clin Genet, 96(1), 43-52.
- Zhang Q*, Yang X*, Wang J*, Li J*, Wu Q, Wen Y, Zhao Y, Zhang X, Yao H, Wu X, Yu S, Wei L#, Bao X# (2019). Genomic mosaicism in the pathogenesis and inheritance of a Rett syndrome Genet Med, 21(6), 1330-1338.
- Liu A#, Yang X#, Yang X, Wu Q, Zhang J, Sun D, Yang Z, Jiang Y, Wu X, Wei L, Zhang Y (2019). Mosaicism and incomplete penetrance of PCDH19 mutations. J Med Genet, 56(2), 81-88.
- Huang AY*,#, Yang X*, Wang S, Zheng X, Wu Q, Ye AY, Wei L# (2018). Distinctive types of postzygotic single-nucleotide mosaicisms in healthy individuals revealed by genome-wide profiling of multiple organs. PLoS Genet, 14(5), e1007395.
- Yang X*, Liu A*, Xu X, Yang X, Zeng Q, Ye AY, Yu Z, Wang S, Huang AY, Wu X, Wu Q#, Wei L#, Zhang Y# (2017). Genomic mosaicism in paternal sperm and multiple parental tissues in a Dravet syndrome cohort. Sci Rep, 7(1), 15677.
- Xu X*, Yang X*, Wu Q*, Liu A, Yang X, Ye AY, Huang AY, Li J, Wang M, Yu Z, Wang S, Zhang Z, Wu X, Wei L#, Zhang Y# (2015). Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of "de novo" SCN1A Mutations in Children with Dravet Syndrome. Hum Mutat, 36(9), 861-72.
